Researchers Create World’s Largest DNA Origami
For Immediate Release
Researchers from North Carolina State University, Duke University and the University of Copenhagen have created the world’s largest DNA origami, which are nanoscale constructions with applications ranging from biomedical research to nanoelectronics.
“These origami can be customized for use in everything from studying cell behavior to creating templates for the nanofabrication of electronic components,” says Dr. Thom LaBean, an associate professor of materials science and engineering at NC State and senior author of a paper describing the work.
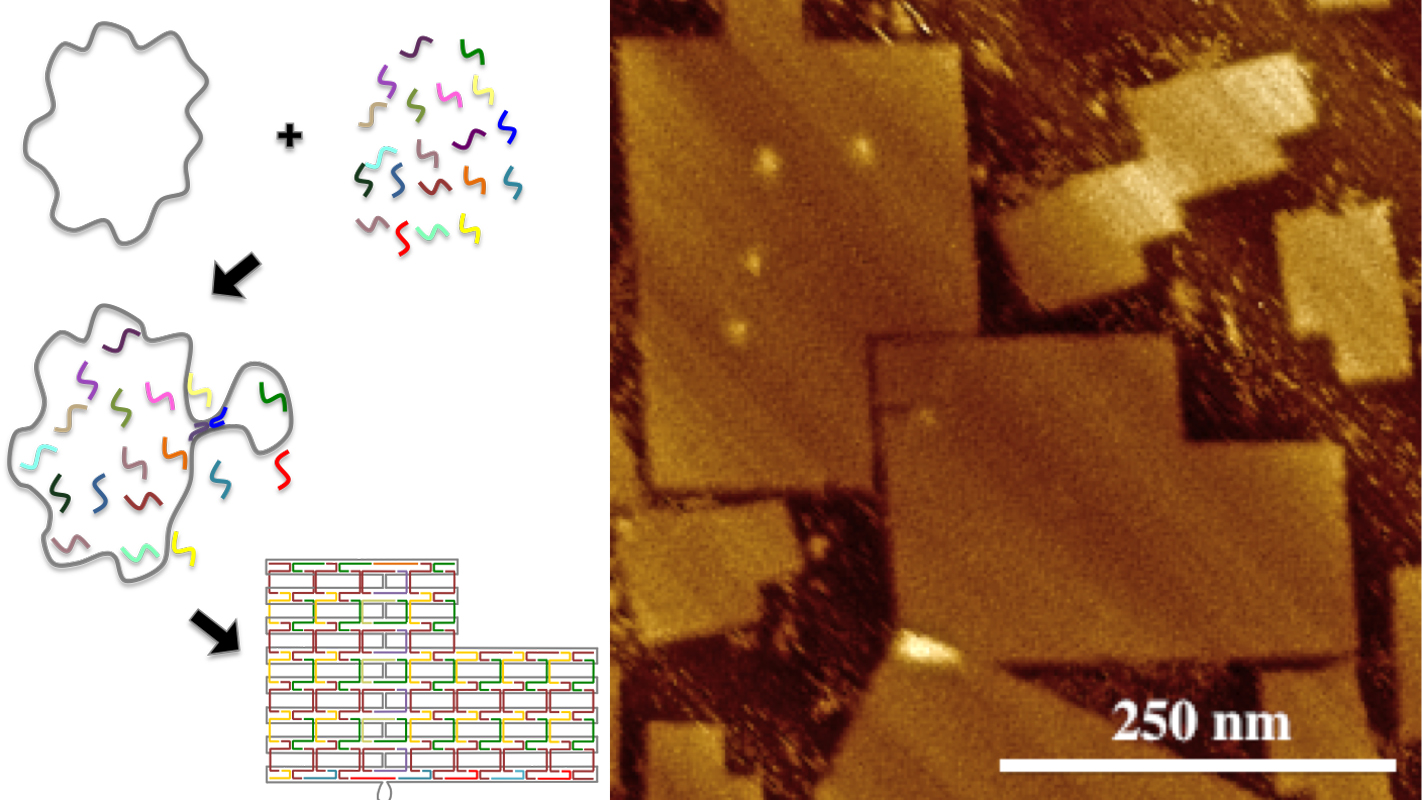
DNA origami are self-assembling biochemical structures that are made up of two types of DNA. To make DNA origami, researchers begin with a biologically derived strand of DNA called the scaffold strand. The researchers then design customized synthetic strands of DNA, called staple strands. Each staple strand is made up of a specific sequence of bases (adenine, cytosine, thaline and guanine – the building blocks of DNA), which is designed to pair with specific subsequences on the scaffold strand.
The staple strands are introduced into a solution containing the scaffold strand, and the solution is then heated and cooled. During this process, each staple strand attaches to specific sections of the scaffold strand, pulling those sections together and folding the scaffold strand into a specific shape.
The standard for DNA origami has long been limited to a scaffold strand that is made up of 7,249 bases, creating structures that measure roughly 70 nanometers (nm) by 90 nm, though the shapes may vary.
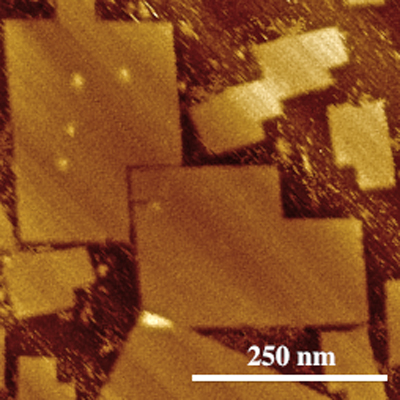
However, the research team led by LaBean has now created DNA origami consisting of 51,466 bases, measuring approximately 200 nm by 300 nm.
“We had to do two things to make this viable,” says Dr. Alexandria Marchi, lead author of the paper and a postdoctoral researcher at Duke. “First we had to develop a custom scaffold strand that contained 51 kilobases. We did that with the help of molecular biologist Stanley Brown at the University of Copenhagen.
“Second, in order to make this economically feasible, we had to find a cost-effective way of synthesizing staple strands – because we went from needing 220 staple strands to needing more than 1,600,” Marchi says.
The researchers did this by using what is essentially a converted inkjet printer to synthesize DNA directly onto a plastic chip.
“The technique we used not only creates large DNA origami, but has a fairly uniform output,” LaBean says. “More than 90 percent of the origami are self-assembling properly.”
The paper, “Toward Larger DNA Origami,” is published online in Nano Letters. The paper was co-authored by Dr. Ishtiaq Saaem, a former Ph.D. student at Duke; Dr. Briana Vogen, a former postdoctoral researcher at NC State; and Dr. Stanley Brown at the University of Copenhagen.
The research was supported by the National Science Foundation under grants CDI-0835794, OISE-1246799, and EPMD-1231888, and by the University of Copenhagen.
-shipman-
Note to Editors: The study abstract follows.
“Toward Larger DNA Origami”
Authors: Alexandria N. Marchi and Ishtiaq Saaem, Duke University; Briana N. Vogen and Thomas H. LaBean, North Carolina State University; Stanley Brown, University of Copenhagen
Published: online Sept. 1, Nano Letters
DOI: 10.1021/nl502626s
Abstract: Structural DNA nanotechnology, and specifically scaffolded DNA origami, is rapidly developing as a versatile method for bottom-up fabrication of novel nanometer-scale materials and devices. However, lengths of conventional single-stranded scaffolds, for example, 7,249-nucleotide circular genomic DNA from the M13mp18 phage, limit the scales of these uniquely addressable structures. Additionally, increasing DNA origami size generates the cost burden of increased staple-strand synthesis. We addressed this 2-fold problem by developing the following methods: (1) production of the largest to-date biologically derived single-stranded scaffold using a λ/M13 hybrid virus to produce a 51 466-nucleotide DNA in a circular, single-stranded form and (2) inexpensive DNA synthesis via an inkjet-printing process on a chip embossed with functionalized micropillars made from cyclic olefin copolymer. We have experimentally demonstrated very efficient assembly of a 51-kilobasepair origami from the λ/M13 hybrid scaffold folded by chip-derived staple strands. In addition, we have demonstrated two-dimensional, asymmetric origami sheets with controlled global curvature such that they land on a substrate in predictable orientations that have been verified by atomic force microscopy.
- Categories:


