How DNA ‘Proofreader’ Proteins Pick and Edit Their Reading Material
For Immediate Release
Researchers from North Carolina State University and the University of North Carolina at Chapel Hill have discovered how two important proofreader proteins know where to look for errors during DNA replication and how they work together to signal the body’s repair mechanism.
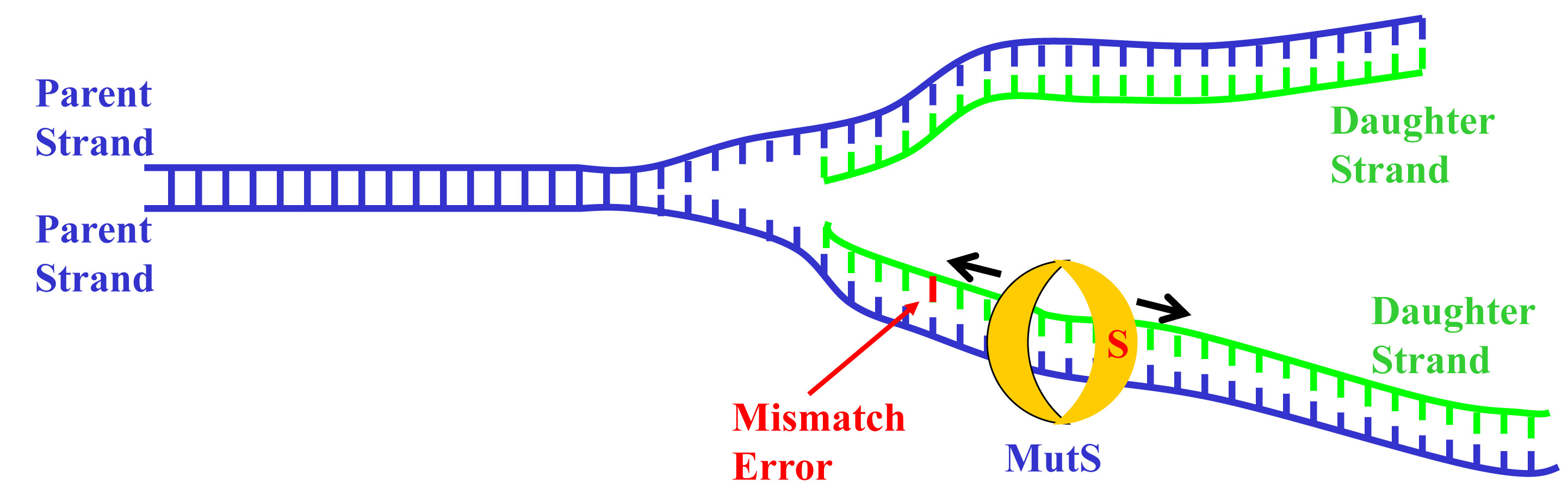
When a cell prepares to divide, the DNA splits first, the double helix “unzipping” into two separate backbones. New nucleotides – adenine, cytosine, guanine or thymine – are filled into the gaps on the other side of the backbone, pairing with their counterparts (adenine with thymine and cytosine with guanine) and replicating the DNA to make a copy for both the old and the new cells. The nucleotides are a correct match most of the time, but occasionally – about one time in a million – there is a mismatch.
Fortunately, our bodies have a system for detecting and repairing these mismatches – a pair of proteins known as MutS and MutL. MutS slides along the newly created side of the DNA strand after it’s replicated, proofreading it. When it finds a mismatch, it locks into place at the site of the error and recruits MutL to come and join it. MutL puts a nick in the newly synthesized DNA strand to mark it as defective and signals a different protein to gobble up the portion of the DNA containing the error. Then the nucleotide matching starts over, filling the gap again. The entire process reduces replication errors around a thousand fold, serving as our body’s best defense against genetic mutations and the problems that can arise from them, like cancer.
“Every living thing uses this process for DNA replication and repair, and we know what the components are, but there are some key mechanisms that aren’t well understood,” says Keith Weninger, professor of physics at NC State and co-author of a paper describing the research. “For example, when the DNA is repaired, it only happens on the new portion of the DNA strand, and not the older template. And the proteins know which way to move along the strand in order to get rid of the defective portion. There’s communication happening here, but disagreement about how it works.”
Scientists know that a protein called PCNA plays an important role in this communication. PCNA sits at the site of a DNA split, and its physical orientation at that site indicates which part of a DNA strand is new. This is called a strand discrimination signal. Somehow, MutS and MutL interact with that signal, allowing MutL to nick the strand in the proper place and indicate need for repair. Scientists have three models for this communication: MutS and MutL combine to form a clamp that slides toward PCNA to interact with the signal, marking the area they slide along for deletion; or they make a fold in the DNA and “pull” the PCNA toward them to the same effect; or MutL somehow coats the defective area, signaling for repair that way.
Weninger, along with co-author Dorothy Erie from UNC-Chapel Hill, set out to determine the most likely method of communication. Using single molecule fluorescence methods, which allowed the researchers to watch one protein moving on one piece of DNA at a time, they determined that when MutS finds an error, it changes shape in a way that allows MutL to bind with it, holding it in place at the site of the mismatch. MutL then changes its shape to “grab” another MutL, and so on, coating the defective part of the strand and nicking the area that needs repair. In other words, Weninger, Erie and their colleagues found that the third model was the correct one.
“This system is essential to keeping the genome stable,” says Weninger. “We know all the players and the beginning and end of the process, but this work fills in one of the middle steps that was completely unknown and very different from what some have imagined.
“We also know that mutations in MutS and MutL are associated with very specific cancers,” he continues. “But if we’re going to figure out how defects lead to disease, we first must understand how these proteins normally do their job. This research is another step toward that understanding.”
The research appears online in Proceedings of the National Academy of Sciences. Funding was provided by the American Cancer Society, the National Institutes of Health and the National Science Foundation. Paul Modrich of Duke University, Manju Hingorani of Wesleyan University, and postdocs and graduate students Ruoyi Qiu (NC State), Elizabeth Sacho (NC State), Hunter Wilkins (UNC-CH), Xindgong Zhang (Duke), and Miho Sakato (Wesleyan) also contributed to the work.
-peake-
Note to editors: Abstract follows
“MutL traps MutS at a DNA mismatch”
DOI: 10.1073/pnas.1505655112
Authors: Keith Weninger, Ruoyi Qiu, Elizabeth Sacho, NC State University; Dorothy Erie, Hunter Wilkins, UNC-Chapel Hill; Paul Modrich, Xingdong Zhang, Duke University; Manju Hingorani, Miho Sakato, Wesleyan University
Published: Online in Proceedings of the National Academy of Sciences
Abstract:
DNA mismatch repair (MMR) identifies and corrects errors made during replication. In all organisms except those expressing MutH, interactions between a DNA mismatch, MutS, MutL, and the replication processivity factor (β-clamp or PCNA) activate the latent MutL endonuclease to nick the error-containing daughter strand. This nick provides an entry point for downstream repair proteins. Despite the well-established significance of strand-specific nicking in MMR, the mechanism(s) by which MutS and MutL assemble on mismatch DNA to allow the subsequent activation of MutL’s endonuclease activity by β-clamp/PCNA remains elusive. In both prokaryotes and eukaryotes, MutS homologs undergo conformational changes to a mobile clamp state that can move away from the mismatch. However, the function of this MutS mobile clamp is unknown. Furthermore, whether the interaction with MutL leads to a mobile MutS–MutL complex or a mismatch-localized complex is hotly debated. We used single molecule FRET to
determine that Thermus aquaticus MutL traps MutS at a DNA mismatch after recognition but before its conversion to a sliding clamp. Rather than a clamp, a conformationally dynamic protein assembly typically containing more MutL than MutS is formed at the mismatch. This complex provides a local marker where interaction with β-clamp/PCNA could distinguish parent/daughter strand identity. Our finding that MutL fundamentally changes MutS actions following mismatch detection reframes current thinking on MMR signaling processes critical for genomic stability.
- Categories:


