Salty Algae Leads to Sweet Success
What do pink flamingos, a biofuels patent application and a recent paper in the esteemed scientific journal Nature have in common?
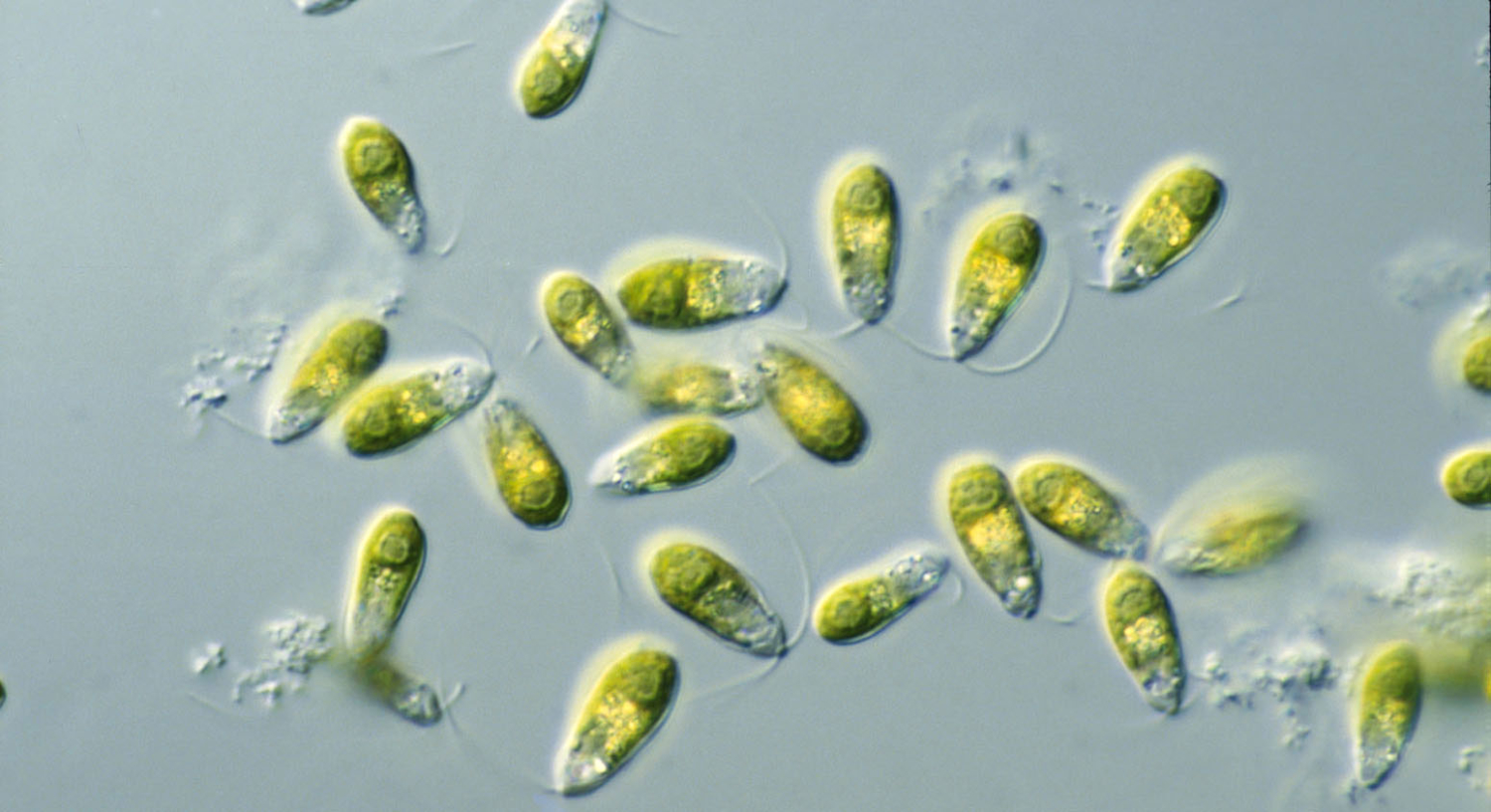
Dunaliella, a group of saltwater algae!
Algae in this group produce significant amounts of orange-pink pigments, and when flamingos feed on the algae — and the tiny shrimp that also eat the algae — the pigments are absorbed by the birds.
Heike Sederoff, a professor in the Department of Plant and Microbial Biology, started studying these algae as part of a project to make biofuels from algae. This research led to a biofuels patent application and a recent paper in the journal Nature.
 The paper shares the results of a large, international collaboration to study how plants evolved. The plant scientists compared the genes actively produced by 1,124 different species of plants from algae and ferns to pine trees and rice. The international consortium of almost 200 plant scientists is called the One Thousand Plant Transcriptomes Initiative, and is led by James H. Leebens-Mack at the University of Georgia and Gane Ka-Shu Wong at the University of Alberta.
The paper shares the results of a large, international collaboration to study how plants evolved. The plant scientists compared the genes actively produced by 1,124 different species of plants from algae and ferns to pine trees and rice. The international consortium of almost 200 plant scientists is called the One Thousand Plant Transcriptomes Initiative, and is led by James H. Leebens-Mack at the University of Georgia and Gane Ka-Shu Wong at the University of Alberta.
Sederoff was introduced to the initiative while attending a scientific conference in Singapore.
“This was a big, international community effort where we pooled all this information about the different species and classes of plants to get, fundamentally, the plant family tree,” Sederoff said. “We contributed the saltwater algae transcriptomes, while a small team was involved in the heavy lifting of doing the computational analysis.”
The initiative analyzed the plants’ transcriptome, or the set of genes actively turned into RNA instead of the plants’ genome, or the complete set of genes for an organism, because it is cheaper, easier and faster, Sederoff said. Plants can have huge genomes with many similar genes, and the transcriptome provides enough information to gain new insights into how plants evolved and are related to each other.
The initiative found that the duplication of whole plant genomes or portions of the genome is fairly common, and evolutionarily advantageous. These duplication events give plants more genetic tools for adapting to new challenges and evolving major innovations such as vasculature — one key difference between mosses and ferns — and flowers — one key difference between pine trees and cherry trees.
If a plant only has one copy of a gene, a mutation in that gene could prove deadly, Sederoff said. However, if a plant has multiple copies of a gene, one copy can maintain the original function while the other is free to “play around” and potentially produce new functions.
“Plants are everywhere; everyone sees them, eats them and uses them — for paper, furniture and clothing,” Sederoff said. “And understanding where plants came from, how they are related, how they evolved, from green algae to trees, is very cool.”
Making biofuels with algae
Sederoff began working with the saltwater algae more than ten years ago as part of a National Science Foundation (NSF) project to turn algae into renewable biofuel to replace fossil fuels. The NSF project was led by Bill Roberts, a professor of mechanical and aerospace engineering. As the goal was to go from growing algae that produce large quantities of oil and extracting that oil to turning it into fuel and testing it in combustion engines, it was a truly interdisciplinary project involving both engineers and biologists.
For that project it was essential to start with saltwater algae, in order to not consume precious fresh water.
When the team began the project, they thought getting algae would be easy, Sederoff said. However, the samples they ordered were not a collection of genetically identical algae — which is critical for solid research — so they needed to sequence the algae’s transcriptome.
“The Nature paper was a little side benefit from having the algae transcriptomes for the biofuels research,” Sederoff said.
Another outcome from having the algae transcriptomes, was that Sederoff and Amy Grunden, a William Neal Reynolds Distinguished Professor in the Department Plant and Microbial Biology, were able to get funding from the Chancellor’s Innovation Fund to engineer bacterial genes for producing economically-valuable enzymes into the algae.
This is because one common challenge with using algae to produce biofuels, is ensuring it is cost effective. Sederoff said, “Because petroleum is so cheap, you need to make something else too, a co-product, to offset the costs.”
The team’s patent application focuses on using the saltwater algae to produce extremozymes — enzymes that work under extreme conditions and are quite useful for industry, and ordinary consumers, such as in the form of stain-fighting and grease-gobbling laundry detergent. The extremozymes they studied include enzymes used for making cheese and those that can degrade sarin — an extremely toxic nerve gas.
And because, unlike plants, the algae don’t have cell walls, it is easy to extract the oil for biofuel and the extremozymes from the algae.
However, the algae tended to delete the new genes for producing the valuable extremozymes, she said. So, her research team eventually turned to a canola-like plant that can grow on poor land with little water for their biofuel production research.
They are engineering Camelina sativa plants to produce more oil while requiring less water and fertilizer. Her research team is also exploring ways to improve the efficiency of carbon dioxide fixation. This ongoing research is supported by the U.S. Department of Energy.
Our research addresses grand challenges — and overcomes them.
This post was originally published in College of Agriculture and Life Sciences News.
- Categories:


