Hands-On Model Helps Students Understand Genetic Engineering
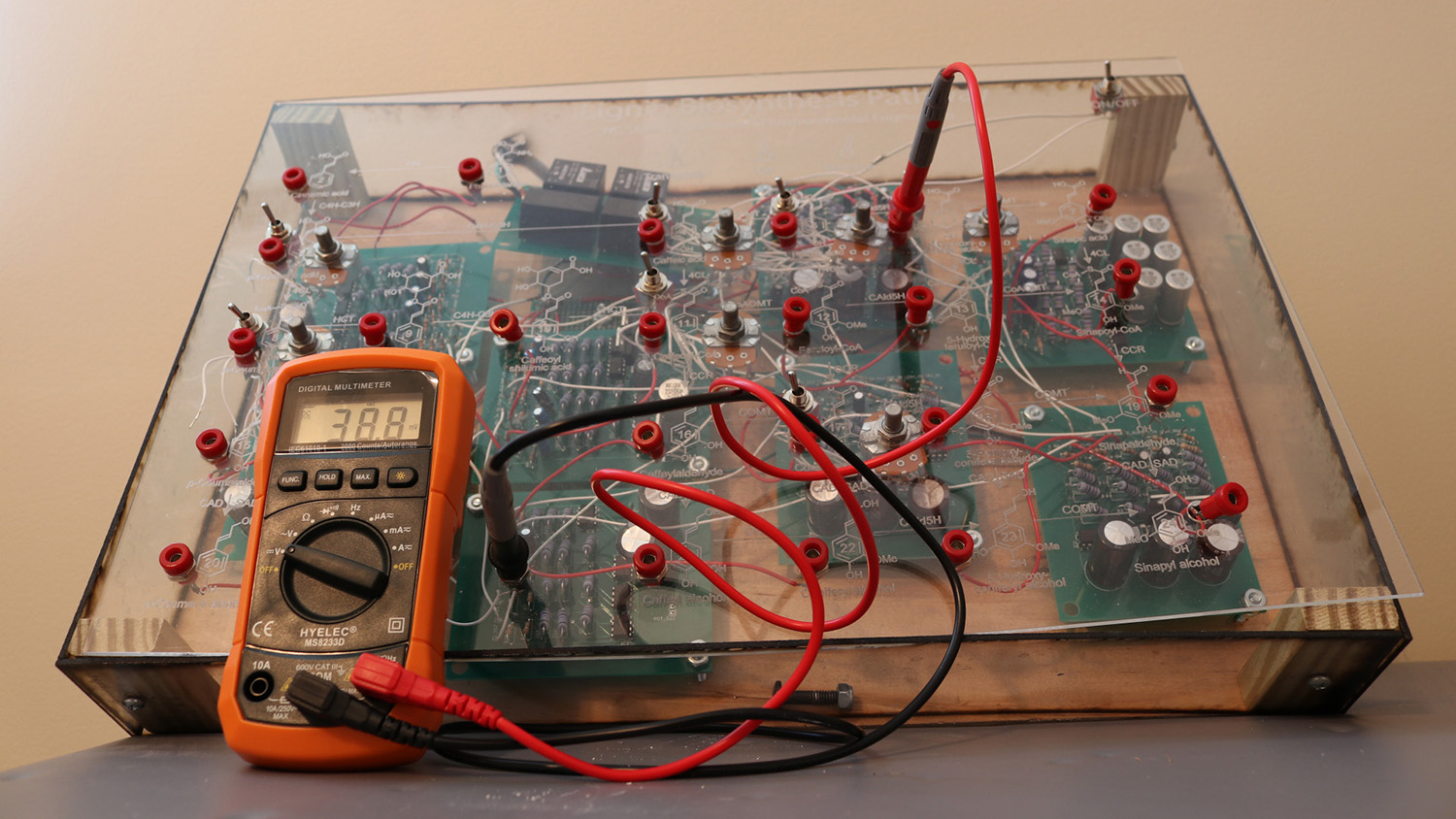
A team of faculty and undergraduate researchers have created a physical, interactive model to teach students about genetics and biological processes at the cellular level.
“By creating an analogue between a biological system and a physically engineered system, we think we can help students grasp the concepts behind genetics, cellular metabolism and genetic engineering,” says Cranos Williams, an expert on biological systems modeling and associate professor of electrical and computer engineering (ECE) at NC State.
The researchers are seeking partners in nonprofit or private sectors to make the tool more widely available for use in K-12 and undergraduate classrooms.
The Big Idea

“Years ago, Cranos and I worked on a project to create a mathematical model of the biological processes plants use to create lignin – which is a substance in plant matter that poses problems for biofuels and paper manufacturing,” says Joel Ducoste, a modeling expert and professor of civil, construction and environmental engineering at NC State.
“We recognized that it is difficult for people to understand what’s happening in a mathematical model, so we wanted to see if it would be possible to create a physical model in which electronic circuits mimic what’s happening in cellular metabolism. That was the genesis of the project.”
To that end, in 2014 Ducoste and Williams enlisted the help of Reuben Valeriano, an ECE undergraduate student at NC State. A year later, they brought on another ECE undergrad, Cody Ellington. By autumn 2016, the team had developed a fully functional prototype of the physical model, called the Enzyme Metabolite Circuit (EzMC).
How It Works
“In a cell, metabolism is a series of chemical reactions that convert one form of mass into another form of mass, through different biochemical pathways,” Williams says. “These reactions can be sequential; they can happen in parallel, with multiple reactions occurring simultaneously; they can trigger additional reactions; or they can occur almost independently. These reactions are how plants convert water, light and whatever is available in the soil into the things that plants need – such as the lignin found in wood.”
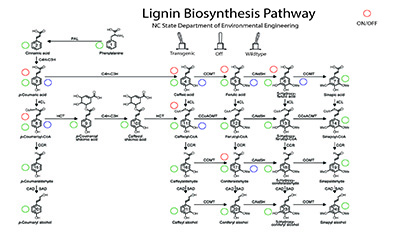
In the EzMC, electricity stands in for mass. For example, in the EzMC, current stands in for mass flux in a cell’s metabolism: the higher the rate at which mass is converted from Material A to Material B in a cell, the higher the current is between two analogous points in the model.
By the same token, the amount of mass being conserved in one form in the cell is represented as voltage (i.e., the higher the amount of mass being stored at one point in the metabolic pathway, the higher the amount of voltage that is stored at that point in the EzMC).
“In our prototype, you can track what is happening in the model by touching a multimeter at various probe points in the model,” Valeriano says. “The multimeter measures voltage, so it effectively tells you how much ‘mass’ is found at any point in the metabolic pathway physical model.
“With additional funding, we could replace those probe points with LED panels that would show how voltage changes at every junction whenever the system is modified – or we could replace the probe points with a computer readout of changes in the metabolic model,” Valeriano says.
Educational Utility
The prototype allows users to make changes at any point in the metabolic pathway – akin to making cellular changes through genetic engineering. Users can then observe how those changes affect cellular behavior.
“We want to convey the idea of what happens when you make genetic modifications to an organism,” Williams says. “Cellular systems are incredibly complex: making one small change may alter behavior in a broad suite of systems – or you may need to make multiple changes in order to alter behavior at all. These are fundamental ideas that we want students to understand. And we think the EzMC could help advance that understanding.”
- Categories:


