Researchers Show What Drives a Novel, Ordered Assembly of Alternating Peptides
For Immediate Release
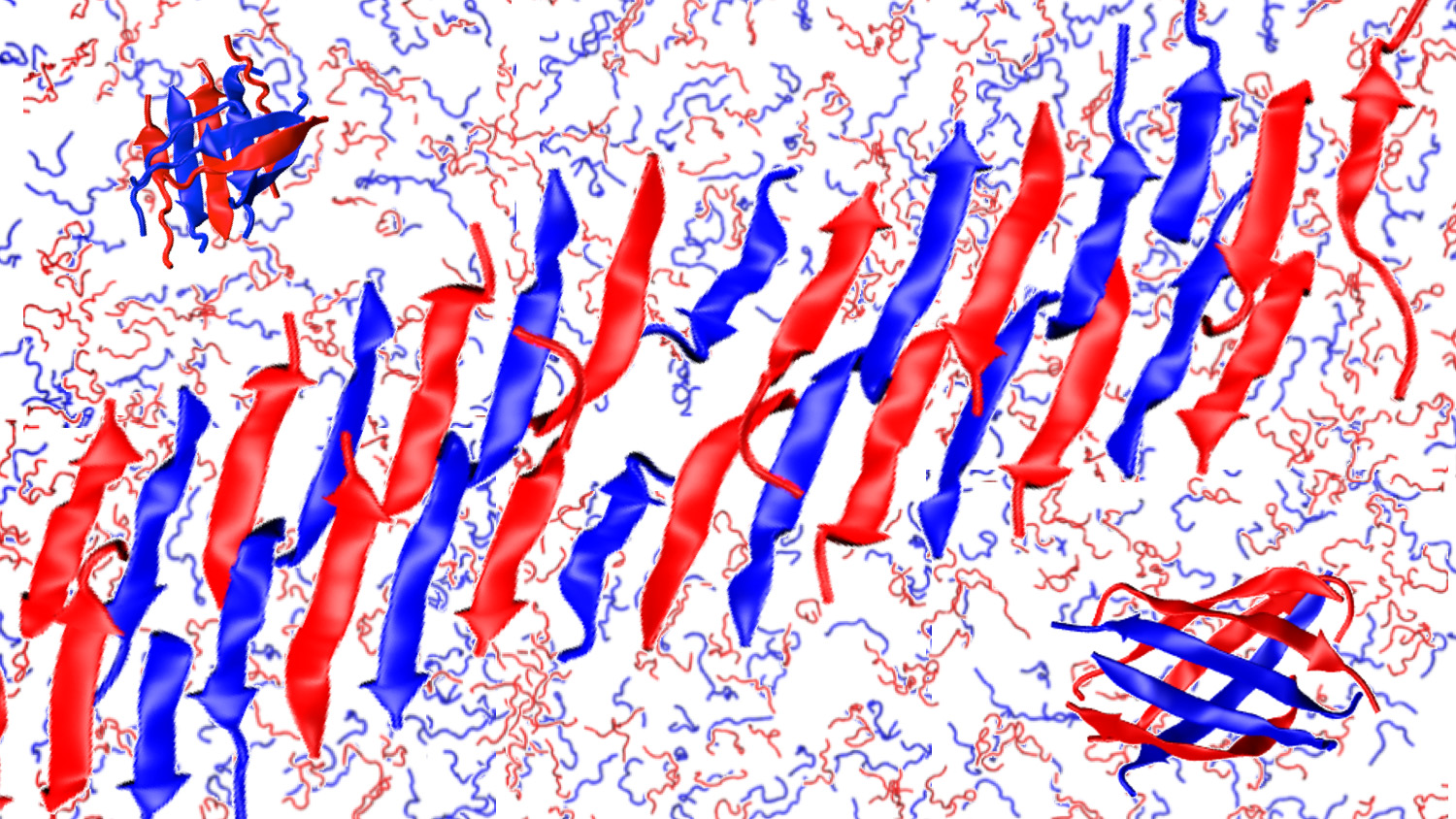
A team of researchers has verified that it is possible to engineer two-layered nanofibers consisting of an ordered row of alternating peptides, and has also determined what makes these peptides automatically assemble into this pattern. The fundamental discovery raises the possibility of creating tailored “ABAB” peptide nanofibers with a variety of biomedical applications.
Peptides are small proteins, made up of short strands of amino acids. It’s well established that peptides can self-assemble into nanofibers composed of beta-sheets. However, that self-assembly normally involves identical copies of the same molecule – molecule A connects to another molecule A.
The new work proves not only that alternating peptides can create these beta sheets – in an ABAB pattern – but why it happens.
“Our team drew on computational simulations, nuclear magnetic resonance (NMR) observations and experimental approaches for this work, and we now know what drives the creation of these alternating peptide structures,” says Carol Hall, corresponding author of a paper on the work and Camille Dreyfus Distinguished University Professor of Chemical and Biomolecular Engineering at North Carolina State University.
“This is important because once you understand why peptides in these ABAB structures are behaving in this way, you can develop more of them,” Hall says.
For this study, researchers worked with a pair of peptides called CATCH(+) and CATCH(-). When introduced into a solution, the peptides array themselves in a row, alternating the two peptides. The peptides also assemble in two beta-sheet layers per nanofiber.
The study itself involved three components. Greg Hudalla’s lab at the University of Florida created the peptides, facilitated the co-assembly of the peptide beta sheets and performed experimental work that provided an overview of the system and its behavior. Hudalla co-authored the paper and is an associate professor in UF’s J. Crayton Pruitt Family Department of Biomedical Engineering.
Meanwhile, Anant Paravastu’s team at Georgia Tech used solid-state NMR to measure the precise relative positions of atoms and molecules in the ABAB peptide beta-sheets. Paravastu co-authored the paper and is an associate professor in Georgia Tech’s School of Chemical and Biomolecular Engineering.
Lastly, Hall’s team at NC State conducted computational simulations to determine what was driving the behavior seen by the researchers at UF and Georgia Tech.
There appear to be multiple forces at play in guiding the assembly of the alternating peptide structures. One of the two types of peptide is negatively charged, while the second type is positively charged. Because positive and negative attract each other, while peptides of the same charge repel each other, this leads to the alternating order of peptides in the strand.
Another aspect of the system’s organization, the stacking, is driven by the types of amino acids in each peptide. Specifically, some of the amino acids in each peptide are hydrophobic, while others are hydrophilic. The hydrophobic amino acids, in effect, want to stick to each other, which results in the two-layer “stacking” effect seen in the beta-sheets.
“It is important that different forces balance to produce the target structure,” Hall says. “If any one of the molecular forces is too strong or too weak, the molecules may never dissolve in water or may fail to recognize their intended partners. Rather than an ordered nanostructure, the molecules could form a disorganized mess, or no structure at all.”
“We’re interested in this because it gives us a glimpse into the fundamental nature of how these systems can work,” Hudalla says. “We’re not aware of any similar co-assembling systems in nature that resemble the system we’ve made here.
“Co-assembling peptide systems hold promise for biomedical applications because we can attach proteins to the A or B peptides that have some specific utility. For example, we could create a peptide scaffold that holds a regular array of enzymes, and those enzymes could serve as catalysts for influencing body chemistry in localized areas.”
“The structures we’re making here are impressive, but they are still not as precise and complex as biological structures that we see in nature,” Paravastu says. “By the same token, we’re not aware of natural structures that contain this alternating peptide structure. This is a good start. We are excited to see where it goes.”
“This work would not have been possible without drawing on the diverse areas of expertise in this research group,” Hall says.
The paper, “Anatomy of a selectively coassembled β-sheet peptide nanofiber,” is published in the journal Proceedings of the National Academy of Sciences. First authors of the paper were Qing Shao, a former postdoctoral researcher at NC State; Kong Wong of Georgia Tech; and Dillon Seroski of UF. The paper was co-authored by Yiming Wang, a former Ph.D. student at NC State; and Renjie Liu of UF.
The work was done with support from the National Science Foundation under grant number 1743432.
-shipman-
Note to Editors: The study abstract follows.
“Anatomy of a selectively coassembled β-sheet peptide nanofiber”
Authors: Qing Shao, Yiming Wang and Carol K. Hall, North Carolina State University; Kong M. Wong and Anant K. Paravastu, Georgia Institute of Technology; Dillon T. Seroski, Renjie Liu and Gregory A. Hudalla, University of Florida
Published: Feb. 18, Proceedings of the National Academy of Sciences
DOI: 10.1073/pnas.1912810117
Abstract: Peptide self-assembly, wherein molecule A associates with other A molecules to form fibrillar β-sheet structures, is common in nature and widely used to fabricate synthetic biomaterials. Selective coassembly of peptide pairs A and B with complementary partial charges is gaining interest due to its potential for expanding the form and function of biomaterials that can be realized. It has been hypothesized that charge-complementary peptides organize into alternating ABAB-type arrangements within assembled β-sheets, but no direct molecular-level evidence exists to support this interpretation. We report a computational and experimental approach to characterize molecular-level organization of the established peptide pair, CATCH. Discontinuous molecular dynamics simulations predict that CATCH(+) and CATCH(−) peptides coassemble but do not self-assemble. Two-layer β-sheet amyloid structures predominate, but off-pathway β-barrel oligomers are also predicted. At low concentration, transmission electron microscopy and dynamic light scattering identified nonfibrillar ∼20-nm oligomers, while at high concentrations elongated fibers predominated. Thioflavin T fluorimetry estimates rapid and near-stoichiometric coassembly of CATCH(+) and CATCH(−) at concentrations ≥100 μM. Natural abundance 13C NMR and isotope-edited Fourier transform infrared spectroscopy indicate that CATCH(+) and CATCH(−) coassemble into two-component nanofibers instead of self-sorting. However, 13C–13C dipolar recoupling solid-state NMR measurements also identify nonnegligible AA and BB interactions among a majority of AB pairs. Collectively, these results demonstrate that strictly alternating arrangements of β-strands predominate in coassembled CATCH structures, but deviations from perfect alternation occur. Off-pathway β-barrel oligomers are also suggested to occur in coassembled β-strand peptide systems.


