Study Could Lead to Power Over ‘Man Eater’

Scientists have long had a name for a gruesome insect that feeds on live flesh of warm-blooded mammals: C. hominivorax, Latin for “man eater.” But now, they have the parasite’s number.
In a paper published this week in the journal Communications Biology, researchers from across the United States and beyond describe the assembly and analysis of the New World screwworm’s genome — a map of the fly’s 534 million DNA base pairs.
Max Scott, a North Carolina State University professor of entomology, is the lead author of the study. He said the research not only sheds light on the insect’s biology and evolution, it also identifies genes that could be targeted to develop control measures that are less expensive than the sterile insect technique that now keeps the insect out of North America.
A threat to livestock, wildlife, pets and humans
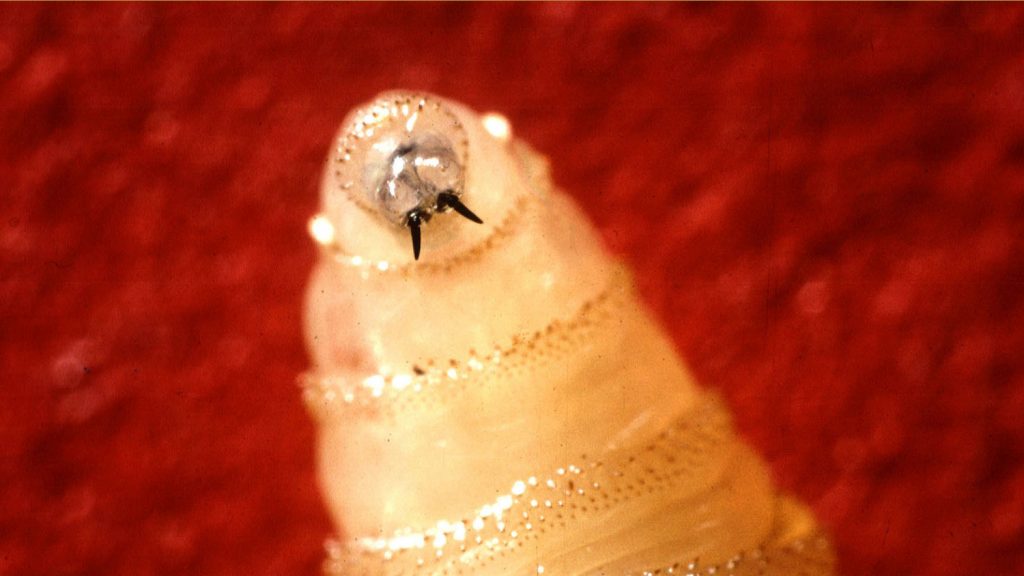
New World screwworms are native to the Western Hemisphere, posing a serious threat to livestock, wildlife, pets and even humans. The flies lay eggs – up to 400 at a time – in wounds and other openings of the skin of warm-blooded mammals.
The fly larvae then bore into the host’s flesh, causing painful lesions as they feed for about five to seven days. Other parasites will feed on tissue that the screwworm kills, and, left untreated, the infections can cause the host to die.
Each year, the screwworm causes billions of dollars in losses for South American and Caribbean farmers.
Effective, but costly, controls

To eradicate the pest from North America, an international insect control program released billions of sterile screwworm flies in the United States, Mexico and Central America over a 50-year period.
A joint program between the United States and Panama has successfully stopped the screwworm from returning by creating a barrier at the border between Panama and Colombia.
The flies prefer to travel over land, so the program saturates the isthmus of Panama with sterile males. About 15 million sterile screwworms are released each week by airplanes over the rainforest that divides the two countries. Wild female screwworms only mate once in their three-week lifetime, and if they mate with a sterile male, they won’t have any offspring.
While the program is effective, feeding millions upon millions of screwworm larvae daily is expensive.
Bringing Down the Cost
Insects’ lifecycles include four stages – embryonic, larval, pupal and adult. Scott said that the cost of producing so many flies could be cut significantly if scientists could figure out how to prevent female offspring entirely or to keep females from maturing beyond the embryonic stage.
Scott and other scientists previously developed a strain of screwworms where females die at the pupal stage, but he said they didn’t have the genetic information they needed to stop female development at earlier stages.
“We needed to identify genes expressed in the embryo, genes that promote cell death and genes that might be involved in sex determination,” Scott said. “We thought it would be a much easier go if we had a whole genome assembly and the genes annotated.
“Along the way,” he added, “we collaborated with other people to see if we could get any insight into the biology of the fly.”
International Team Work Yields Genetic Clues
Scott worked closely on the study with scientists from NC State’s Bioinformatics Research Center, as well as institutions in Ohio, New Mexico, Kentucky, Maryland, Texas, Brazil and Panama. The NC State team did all the DNA and RNA sequencing.
Scientists from the U.S. Department of Agriculture’s Agricultural Research Service stationed in Texas and Panama led efforts to create the highly inbred strain of flies used in the study, and researchers at the National Human Genome Research Institute assembled the genome sequence.
Researchers led by Josh Benoit at the University of Cincinnati conducted what’s known as a developmental gene expression study on RNA sequences from male and female screwworms at different stages in their lifecycle.
[pullquote align=”right” color=”red”]We now have the genes identified that were needed to produce efficient genetic systems for suppression of the pest.[/pullquote]
A group at the University of Kentucky identified genes involved in smell and taste. And scientists from the University of São Paulo, Brazil, identified genes that appear to be evolving quickly and, thus, could be important in the evolution of the fly as an obligate parasite – one that can’t complete its lifecycle without exploiting a host animal.
Scott said the knowledge gained and genetic tools developed through the study have already yielded scientific results. In as-yet-unpublished research, Scott and others have developed strains of New World screwworms where the females don’t survive the embryonic stage.
“With the CRISPR-Cas9 gene editing tools we have developed for screwworm, we are now well placed to determine the importance of specific genes for the screwworms parasitic lifestyle,” Scott said. “We also now have the genes identified that were needed to produce efficient genetic systems for suppression of the pest.”
Abstract: Genomic analyses of a livestock pest, the New World screwworm, find potential targets for genetic control programs
The New World Screwworm fly, Cochliomyia hominivorax, is a major pest of livestock in South America and Caribbean. However, few genomic resources have been available for this species. A genome of 534 Mb was assembled from long read PacBio DNA sequencing of DNA from a highly inbred strain. Analysis of molecular evolution identified 40 genes that are likely under positive selection. Developmental RNA-seq analysis identified specific genes associated with each stage. We identify and analyze the expression of genes that are likely important for host-seeking behavior (chemosensory), development of larvae in open wounds in warm-blooded animals (heat shock protein, immune response) and for building transgenic strains for genetic control programs including gene drive (sex determination, germline). This study will underpin future experiments aimed at understanding the parasitic lifestyle of the screwworm fly and greatly facilitate future development of strains for efficient systems for genetic control of screwworm.
Authors: Maxwell J. Scott1, Joshua B. Benoit2, Rebecca J. Davis1, Samuel Bailey2, Virag Varga2, Ellen O. Martinson3, Paul V. Hickner4, Zainulabeuddin Syed4, Gisele A. Cardoso5, Tatiana T. Torres5, Matthew T. Weirauch6,7,8, Elizabeth H. Scholl9, Adam M. Phillippy10, Agustin Sagel11, Mario Vasquez11, Gladys Quintero11 and Steven R. Skoda12
- Department of Entomology and Plant Pathology, North Carolina State University, Campus Box 7613, Raleigh, NC, 27695-7613
- University of Cincinnati, McMicken School of Arts and Sciences, Dept. of Biological Sciences, Cincinnati, OH, 45221
- Department of Biology, University of New Mexico, Albuquerque, NM, 87131
- Department of Entomology, University of Kentucky, Lexington, KY, 40546
- Department of Genetics and Evolutionary Biology, University of São Paulo, São Paulo, Brazil
- Center for Autoimmune Genomics and Etiology, Cincinnati Children’s Hospital Medical Center, Cincinnati, OH 45229
- Divisions of Biomedical Informatics and Developmental Biology, Cincinnati Children’s Hospital Medical Center, Cincinnati, OH 45229
- Department of Pediatrics, University of Cincinnati College of Medicine, Cincinnati, OH 45267
- Bioinformatics Research Center, North Carolina State University, Campus Box 7566, Raleigh, NC, 27695-7566
- Genome Informatics Section, Computational and Statistical Genomics Branch, National Human Genome Research Institute, Bethesda, MD 20892
- USDA-ARS, Screwworm Research Unit, Pacora, Panama
- USDA-ARS, Tick and Biting Fly Research Unit, Knipling-Bushland Livestock Insects Research Laboratory, 2700 Fredericksburg Rd., Kerrville TX 78028
This post was originally published in College of Agriculture and Life Sciences News.


