Researchers Detail How to Control Shape, Structure of DNA and RNA
For Immediate Release
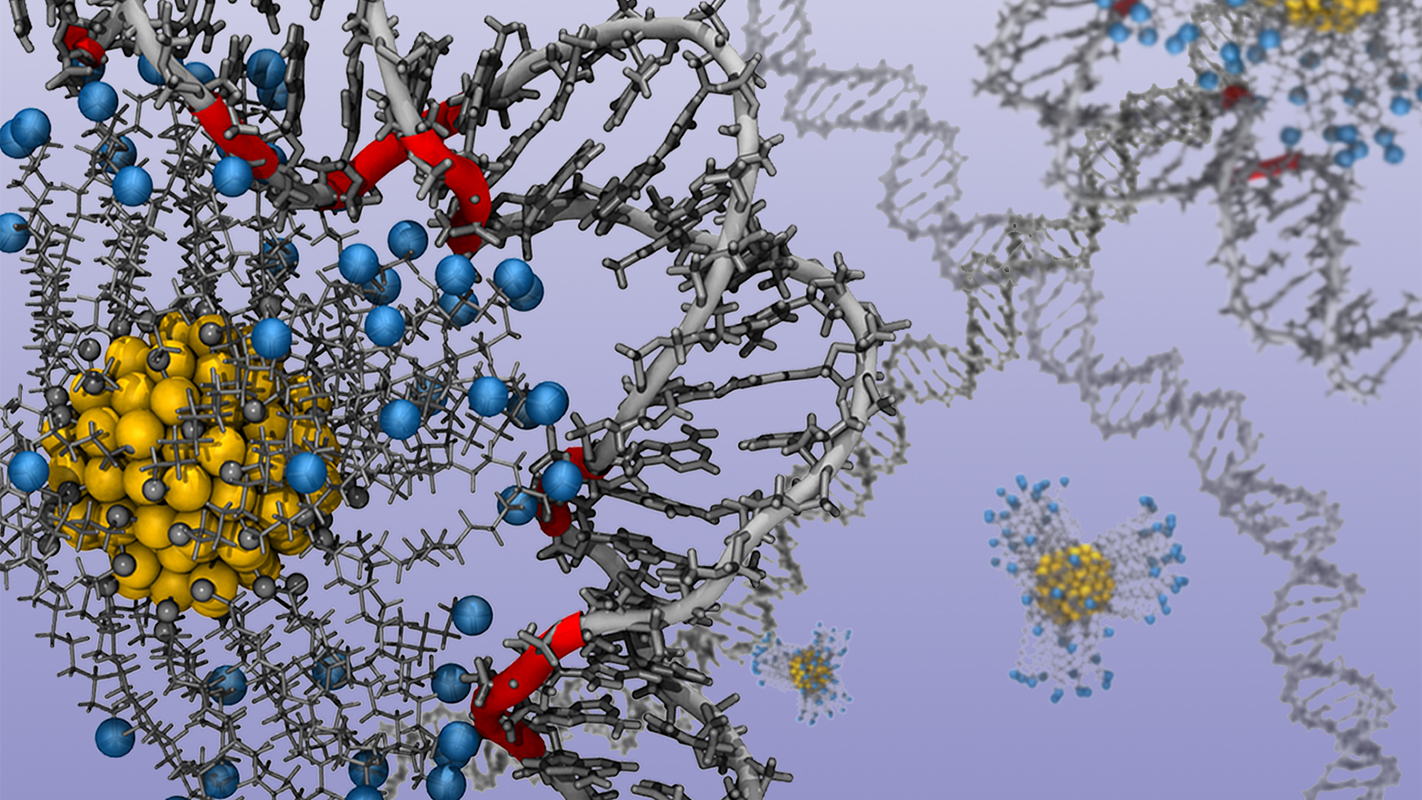
Researchers at North Carolina State University have used computational modelling to shed light on precisely how charged gold nanoparticles influence the structure of DNA and RNA – which may lead to new techniques for manipulating these genetic materials.
The work holds promise for developing applications that can store and transport genetic information, create custom scaffolds for bioelectronics and create new drug delivery technologies.
“In nature, meters of DNA are packed tightly into every living cell,” says Jessica Nash, a Ph.D. student at NC State and lead author of a paper on the work. “This is possible because the DNA is wrapped tightly around a positively charged protein called a histone. We’d like to be able to shape DNA using a similar approach that replaces the histone with a charged gold nanoparticle. So we used computational techniques to determine exactly how different charges influence the curvature of nucleic acids – DNA and RNA.”
In their model, the researchers manipulated the charge of the gold nanoparticles by adding or removing positively charged ligands – organic molecules attached to the surface of the nanoparticle. This allowed them to determine how the nucleic acid responded to each level of charge. An animation of a nanoparticle and ligands shaping a strand of DNA is available at https://www.youtube.com/watch?v=kNpvPyc2bmc&feature=youtu.be.
“This will let researchers know what to expect – how much charge they need in order to get the desired curvature in the nucleic acid,” says Yaroslava Yingling, an associate professor of materials science and engineering at NC State and corresponding author of the paper.
“We used ligands in the model, but there are other ways to manipulate the charge of the nanoparticles,” says Abhishek Singh, a postdoctoral researcher at NC State and co-author of the paper. “For example, if the nanoparticles and nucleic acid are in solution, you can change the charge by changing the pH of the solution.”
The work is also significant because it highlights how far computational research has come in materials science.
“Our large-scale models account for every atom involved in the process,” says Nan Li, a Ph.D. student at NC State and co-author of the paper. “This is an example of how we can use advanced computational hardware, such as the GPUs – or graphics processing units – developed for use in videogames, to conduct state-of-the-art scientific simulations.”
The research team is now building on these findings to design new nanoparticles with different shapes and surface chemistries to get even more control over the shape and structure of nucleic acids.
“No one has come close to matching nature’s efficiency when it comes to wrapping and unwrapping nucleic acids,” Yingling says. “We’re trying to advance our understanding of precisely how that works.”
The paper, “Characterization of Nucleic Acid Compaction with Histone-Mimic Nanoparticles through All-Atom Molecular Dynamics,” is published online in the journal ACS Nano. The paper was co-authored by Abhishek Singh, a postdoctoral researcher at NC State. The work was supported by the National Science Foundation under grants CBET-1403871, DMR-1121107, CMMI-1150682 and DGE-0946818.
-shipman-
Note to Editors: The study abstract follows.
“Characterization of Nucleic Acid Compaction with Histone-Mimic Nanoparticles through All-Atom Molecular Dynamics”
Authors: Jessica A. Nash, Abhishek Singh, Nan K. Li, and Yaroslava G. Yingling, North Carolina State University
Published: Nov. 2, ACS Nano
DOI: 10.1021/acsnano.5b05684
Abstract: The development of nucleic acid (NA) based nanotechnology applications rely on the efficient packaging of DNA and RNA. However, the atomic details of NA-nanoparticle binding remain to be comprehensively characterized. Here, we examined how nanoparticle and solvent properties affect NA compaction. Our large scale all-atom simulations of ligand-functionalized gold nanoparticle (NP) binding to double stranded NAs as a function of NP charge and solution salt concentration reveal different responses of RNA and DNA to cationic NPs. We demonstrate that the ability of a nanoparticle to bend DNA is directly correlated with the NPs charge and ligand corona shape, where more than 50% charge neutralization and spherical shape of the NP ligand corona ensured the DNA compaction. However, NP with 100% charge neutralization is needed to bend DNA almost as efficiently as the histone octamer. For RNA in 0.1 M NaCl, even the most highly charged nanoparticles are not capable of causing bending due to charged ligand end groups binding internally to the major groove of RNA. We show that RNA compaction can only be achieved through a combination of highly charged nanoparticles with low salt concentration. Upon interactions with highly charged NPs, DNA bends through periodic variation in groove widths and depths, whereas RNA bends through expansion of the major groove.
- Categories:


