New Technique Uses Bacteria’s Own CRISPR-Cas System to Turn Off Genes
For Immediate Release
Researchers from North Carolina State University have developed a technique that co-opts an immune system already present in bacteria and archaea to turn off specific genes or sets of genes – creating a powerful tool for future research on genetics and related fields.
“This should not only expedite scientific discovery, but help us better engineer microbial organisms to further biotechnology and medicine,” says Dr. Chase Beisel, an assistant professor of chemical and biomolecular engineering at NC State and senior author of a paper on the work. “For example, this could help us develop bacterial strains that are more efficient at converting plant biomass into liquid fuels.”
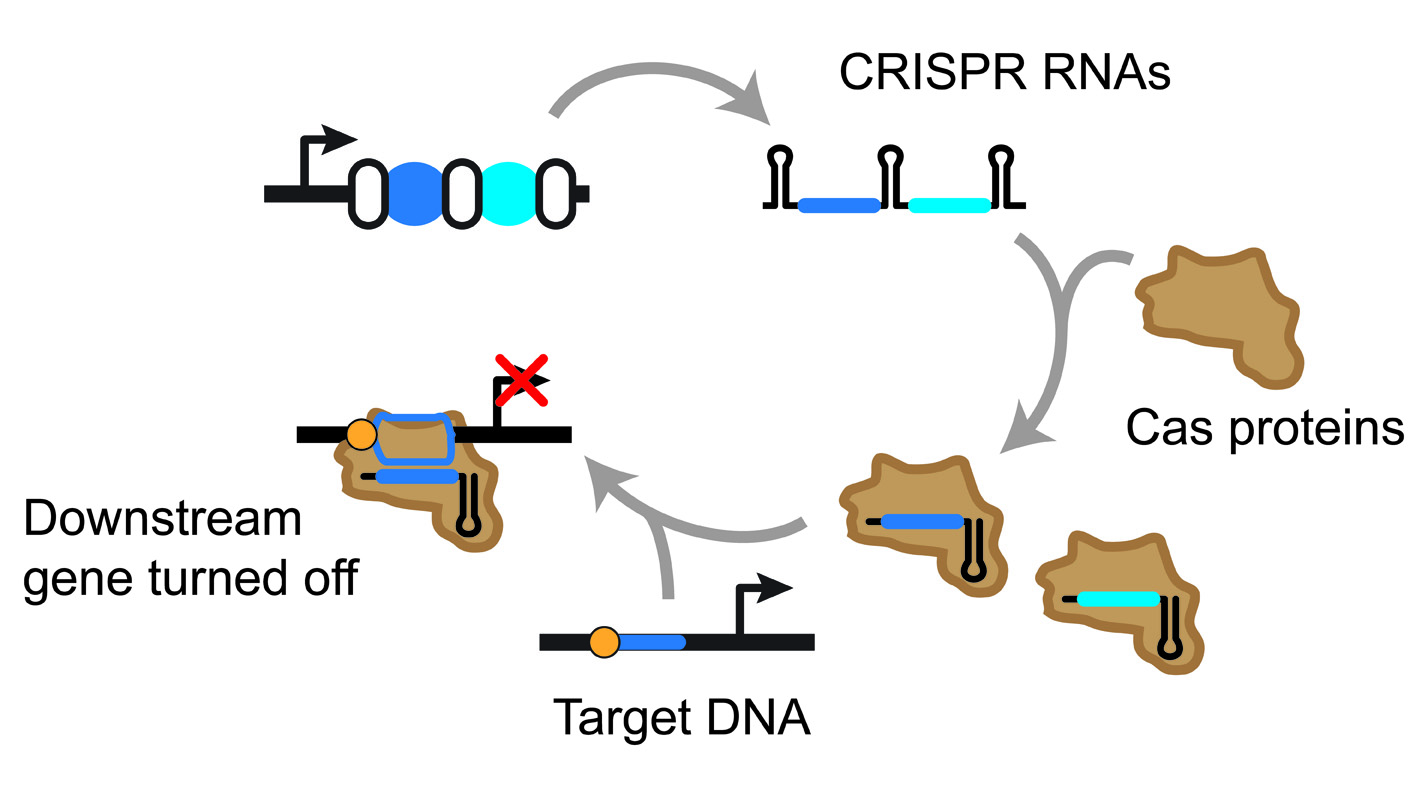
The technique works by hijacking a microbe’s own CRISPR-Cas system. The system normally protects bacteria from invaders such as viruses by creating small strands of RNA called CRISPR RNAs, which match DNA sequences specific to a given invader. In the most common type of CRISPR-Cas system, called a Type I system, a CRISPR RNA and a set of proteins tightly latch onto a matching sequence of DNA. Once bound, the proteins signal for another protein called Cas3 that cuts up the DNA.
“Our approach was to modify the Type I system in two ways,” Beisel says. “First we edit the microbe’s genome to prevent it from producing Cas3, and then we introduce synthetic DNA that produces our customized CRISPR RNAs.”
These customized RNAs include complementary sequences that are tailored to match up with specific DNA sequences in the microbe’s genome.
When a gene is turned on, it transcribes specific RNAs that a cell uses to make proteins. This process is the basis of all biological functions.
But when the customized CRISPR RNAs latch onto a specific gene, those genes are turned off, or blocked from transcribing RNAs. Beisel’s team was able to introduce both individual CRISPR RNAs, and groups of CRISPR RNAs that were chained together, allowing them to target multiple genes at the same time. Some of these genes enable the microbes to consume different sugars. By controlling which genes were turned off, the team could dictate which sugars the microbes could consume.
“This technique will allow researchers to better understand the role of individual genes, or sets of genes, by simply turning those genes off,” Beisel says. “And this approach could be used in conjunction with high-throughput screening techniques to identify gene sets associated with problems such as multidrug resistance or benefits, like those related to probiotics.”
In addition, because the affected genes aren’t destroyed – there are no Cas3 proteins to chop them up – the technique can allow the genes to turn back on. This is done by halting the production of the CRISPR RNAs, which allows affected genes to begin transcribing again as the microbes reproduce and the remaining CRISPR RNA degrades.
The paper, “Repurposing endogenous type I CRISPR-Cas systems for programmable gene repression,” is open access and is published online in the journal Nucleic Acids Research. Lead author of the paper is Michelle Luo, a Ph.D. student at NC State. Co-authors are Adam Mullis, who did the work while an undergraduate at NC State, and Ryan Leenay, a Ph.D. student at NC State. The work was supported the National Science Foundation under grant CBET-1403135 and the National Institutes of Health under grant 5T32GM008776-15.
-shipman-
Note to Editors: The study abstract follows.
“Repurposing endogenous type I CRISPR-Cas systems for programmable gene repression”
Authors: Michelle L. Luo, Adam S. Mullis, Ryan T. Leenay, and Chase L. Beisel, North Carolina State University
Published: online Oct. 17, 2014 in Nucleic Acids Research
DOI: 10.1093/nar/gku971
Abstract: CRISPR-Cas systems have shown tremendous promise as heterologous tools for genome editing and transcriptional regulation. Because these RNA-directed immune systems are found in most prokaryotes, an opportunity exists to harness the endogenous systems as convenient tools in these organisms. Here, we report that the Type I-E CRISPR-Cas system in Escherichia coli can be co-opted for programmable transcriptional repression. We found that deletion of the signature cas3 gene converted this immune system into a programmable gene regulator capable of reversible gene silencing of heterologous and endogenous genes. Targeting promoter regions yielded the strongest repression, whereas targeting coding regions showed consistent strand bias. Furthermore, multi-targeting CRISPR arrays could generate complex phenotypes. This strategy offers a simple approach to convert many endogenous Type I systems into transcriptional regulators, thereby expanding the available toolkit for CRISPR-mediated genetic control while creating new opportunities for genome-wide screens and pathway engineering.
- Categories:


